Google Scholar, ORCID: 0000-0002-4224-798X
Preprints, 2025, 2024, 2023, 2022, 2021, 2020, 2019, 2017, 2016, 2015, 2014, 2013, 2012
Preprints
2026
37. The effect of PEGylation on surface tethering of liposomes via DNA nanotechnology.
J.P.Gaston, S.Meepat, M.S.Islam, J.Li, J.K.D.Singh, M.J.Booth, S.F.J.Wickham, M.A.B.Baker
Journal of Lipid Research, 67, 1, 100940 (2026)
DOI: 10.1016/j.jlr.2025.100940

2025
36. Magnetic Activation of Spherical Nucleic Acids for the Remote Control of Synthetic Cells.
E.Parkes, A.Al Samad, G.Mazzotti, C.Newell, B.Ng, A.Radford, M.J.Booth
Nature Chemistry, 17, 1505 (2025)
DOI: 10.1038/s41557-025-01909-6
(BioRxiv, DOI: 10.1101/2024.08.21.608917)
Research Briefing – The Times article – EUSynCell

35. Strategies and applications of synthetic cell communication.
H.Moghimianavval, C.Newell, P.Parvizian, M.J.Booth, A.Liu
Nature Chemical Biology, 21, 1317 (2025)
DOI: 10.1038/s41589-025-02002-2

34. Harnessing BET-Bromodomain Assisted Nuclear Import for Targeted Subcellular Localization and Enhanced Efficacy of Antisense Oligonucleotides.
D.Kashyap, T.Milne, M.J.Booth
Journal of the American Chemical Society, 147, 32, 29478 (2025)
DOI: 10.1021/jacs.5c09544
(ChemRxiv, DOI: 10.26434/chemrxiv-2024-p6q7p-v2)
NATA article

33. DNA-programmable Protein Degradation: Dynamic Control of PROTAC Activity via DNA Hybridization and Strand Displacement.
D.Kashyap, T.Milne, M.J.Booth
JACS Au, 5, 8, 3799 (2025)
DOI: 10.1021/jacsau.5c00422
(ChemRxiv, DOI: 10.26434/chemrxiv-2025-x4m7p)

32. Engineering antisense oligonucleotides for targeted mRNA degradation through lysosomal trafficking.
D.Kashyap, T.Milne, M.J.Booth
Chemical Science, 16, 13096 (2025)
DOI: 10.1039/D5SC03751D
(ChemRxiv, DOI: 10.26434/chemrxiv-2025-jzgrj)

2024
31. Nucleic Acid Conjugates: Unlocking Therapeutic Potential.
D.Kashyap, M.J.Booth
ACS Bio & Med Chem Au, 5 (1), 3 (2024)
DOI: 10.1021/acsbiomedchemau.4c00092

2023
30. Sculpting DNA-based synthetic cells through phase separation and phase-targeted activity.
L.Malouf, D.A.Tanase, G.Fabrini, R.A.Brady, M.Paez-Perez, A.Leathers, M.J.Booth, L.Di Michele
Chem, 9 (11), 3347 (2023)
DOI: 10.1016/j.chempr.2023.10.004
(BioRxiv, DOI: 10.1101/2023.03.17.533162)

29. Sequence-independent, site-specific incorporation of chemical modifications to generate light-activated plasmids.
K.Chung, M.J.Booth
Chemical Science, 14, 12693 (2023)
DOI: 10.1039/D3SC02761A
(BioRxiv, DOI: 10.1101/2023.05.26.542478)

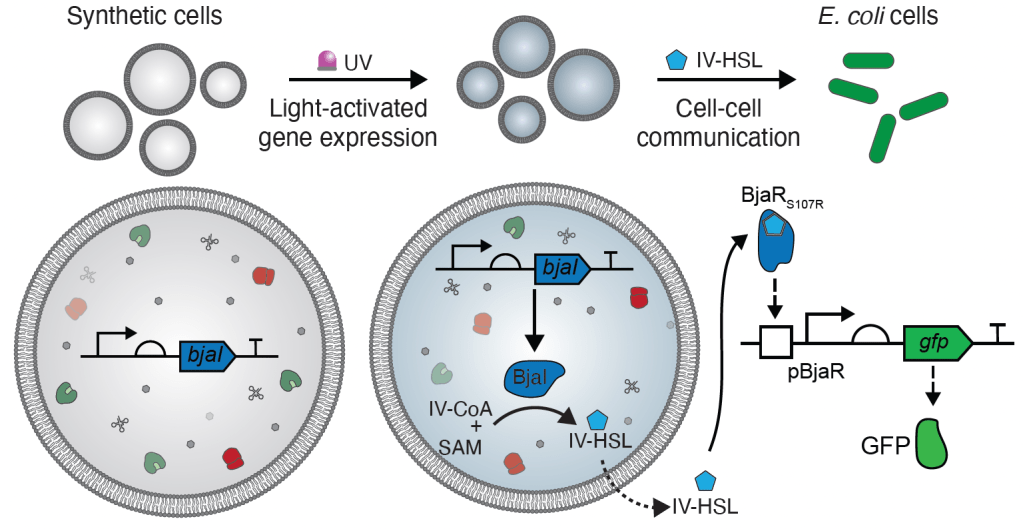
28. Engineering cellular communication between light-activated synthetic cells and bacteria.
J.M.Smith, D.Hartmann, M.J.Booth
Nature Chemical Biology, 19, 1138 (2023)
DOI: 10.1038/s41589-023-01374-7
(BioRxiv, DOI: 10.1101/2022.07.22.500923)
Research Briefing
27. Orthogonal light-activated DNA for patterned biocomputing within synthetic cells.
D.Hartmann, R.Chowdhry, J.M.Smith, M.J.Booth
Journal of the American Chemical Society, 145, 17, 9471 (2023)
DOI: 10.1021/jacs.3c02350
(ChemRxiv, DOI: 10.26434/chemrxiv-2022-p8xgb-v2)
EUSynCell article

26. Precise, orthogonal remote-control of cell-free systems using photocaged nucleic acids.
G.Mazzotti, D.Hartmann, M.J.Booth
Journal of the American Chemical Society, 145, 17, 9481 (2023)
DOI: 10.1021/jacs.3c01238
(ChemRxiv, DOI: 10.26434/chemrxiv-2023-ssv30)

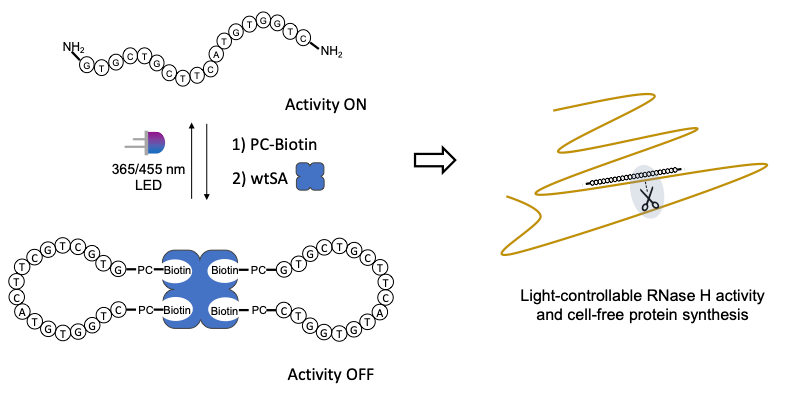
25. Handcuffed antisense oligonucleotides for light-controlled cell-free expression.
D.Hartmann, M.J.Booth
Chemical Communications, 59, 5685 (2023)
DOI: 10.1039/D3CC01374J
24. Accessible light-controlled knockdown of cell-free protein synthesis using phosphorothioate-caged antisense oligonucleotides.
D.Hartmann, M.J.Booth
Communications Chemistry, 6, 59 (2023)
DOI: 10.1038/s42004-023-00860-2
(ChemRxiv, DOI: 10.26434/chemrxiv-2022-1lqc8)

2022
23. DNA and RNA sequencing.
M.J.Booth
Nucleic Acids in Chemistry and Biology: Edition 4 Editors: G Michael Blackburn, Martin Egli, Michael J Gait, Jonathan K Watts. ISBN 978-1-78801-904-0 (2022)

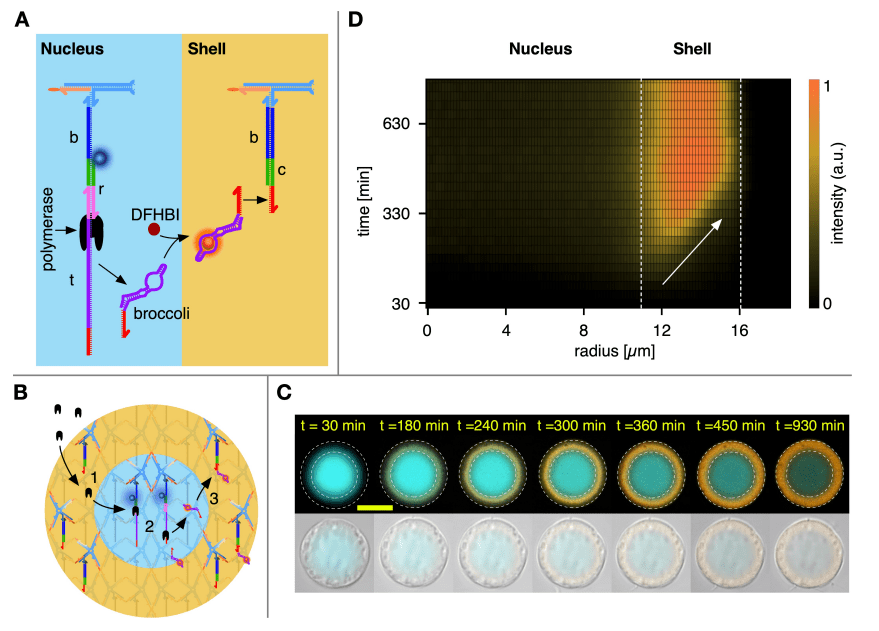
22. Reaction–Diffusion Patterning of DNA-Based Artificial Cells.
A.Leathers, M.Walczak, R.A.Brady, A.Al Samad, J.Kotar, M.J.Booth, P.Cicuta, L.Di Michele
Journal of the American Chemical Society, 144, 38, 17469 (2022)
DOI: 10.1021/jacs.2c06140
(BioRxiv, DOI: 10.1101/2022.03.24.485404)
21. Controlling Synthetic Cell-Cell Communication.
J.M.Smith, R.Chowdhry, M.J.Booth
Frontiers in Molecular Bioscience, 8, 809945 (2022)
DOI: 10.3389/fmolb.2021.809945

2021
20. A Lipid-Based Droplet Processor for Parallel Chemical Signals.
I.Cazimoglu, M.J.Booth, H.Bayley
ACS Nano, 15, 12, 20214 (2021)
DOI: 10.1021/acsnano.1c08217
(BioRxiv, DOI:10.1101/2021.05.05.442835)

19. Reduced Bisulfite Sequencing: Quantitative Base-Resolution Sequencing of 5-Formylcytosine.
M.J.Booth, S.Balasubramanian
Methods in Molecular Biology: TET Proteins and DNA Demethylation, 2272, 3-12 (2021)
DOI:10.1007/978-1-0716-1294-1_1

2020
18. Controlling gene expression with light: a multidisciplinary endeavour.
D.Hartmann, J.M.Smith, G.Mazzotti, R.Chowdhry, M.J.Booth
Biochemical Society Transactions, BST20200014 (2020)
DOI:10.1042/BST20200014

17. Transmembrane protein rotaxanes reveal kinetic traps in the refolding of translocated substrates.
J.Feng, P.Martin-Baniandres, M.J.Booth, G.Veggiani, M.Howarth, H.Bayley, D.Rodriguez-Larrea
Communications Biology, 3, 159 (2020)
DOI:10.1038/s42003-020-0840-5

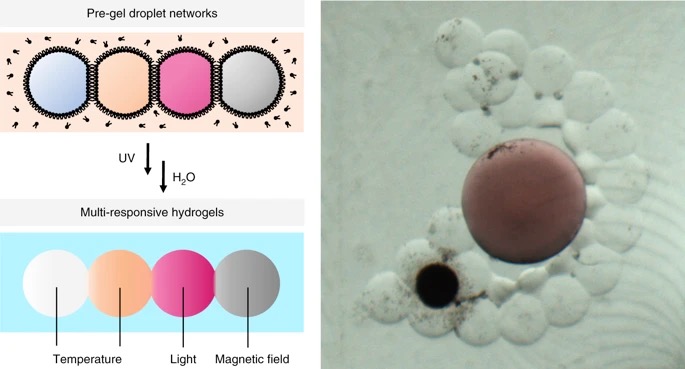
16. Multi-responsive hydrogel structures from patterned droplet networks.
F.G.Downs, D.J.Lunn, M.J.Booth, J.B.Sauer, W.J.Ramsay, R.G.Klemperer, C.J.Hawker, H.Bayley
Nature Chemistry, 12, 363 (2020)
DOI:10.1038/s41557-020-0444-1
2019
15. Controlled deprotection and release of a small molecule from a compartmented synthetic tissue module.
M.J.Booth, I.Cazimoglu, H.Bayley
Communications Chemistry, 2, 142 (2019)
DOI:10.1038/s42004-019-0244-y
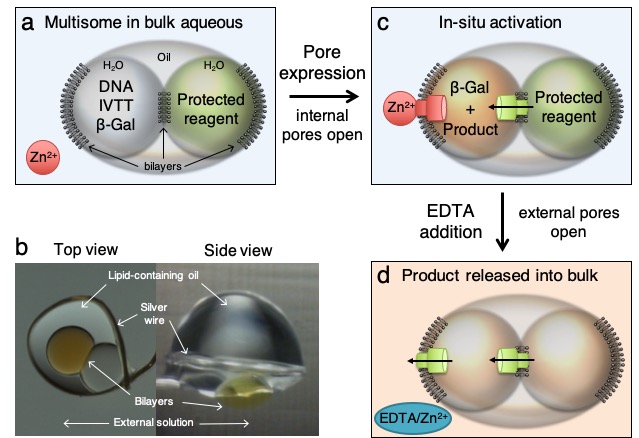
14. Droplet Networks, from Lipid Bilayers to Synthetic Tissues.
M.J.Booth, V.Restrepo-Schild, F.G.Downs, H.Bayley
Encyclopedia of Biophysics (2019)
DOI:10.1007/978-3-642-35943-9_567-1

2017
13. Light-patterning of synthetic tissues with single droplet resolution.
M.J.Booth, V.Restrepo-Schild, S.J.Box, H.Bayley
Scientific Reports, 7, 9315 (2017)
DOI:10.1038/s41598-017-09394-9

12. Functional aqueous droplet networks.
M.J.Booth, V.Restrepo-Schild, F.G.Downs, H.Bayley
Molecular Biosystems, 13, 1658-1691 (2017)
DOI:10.1039/C7MB00192D
11. Light-patterned current generation in a droplet bilayer array.
V.Restrepo-Schild, M.J.Booth, S.J.Box, S.N.Olof, M.Radhakrishnan, H.Bayley
Scientific Reports, 7, 46585 (2017)
DOI:10.1038/srep46585

2016
10. 3D-printed synthetic tissues.
M.J.Booth, H.Bayley
The Biochemist, 38 (4), 16 (2016)
DOI:http://www.biochemist.org/bio/03804/0016/038040016.pdf

9. Light-activated communication in synthetic tissues.
M.J.Booth, V.Restrepo-Schild, A.D.Graham, S.N.Olof, H.Bayley
Science Advances, 2 (4), e1600056 (2016)
DOI:10.1126/sciadv.1600056
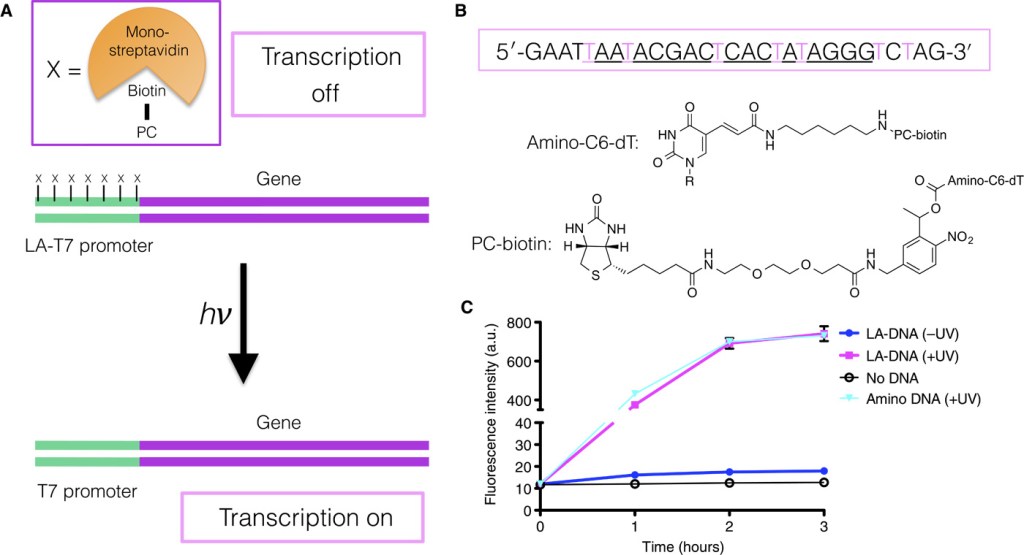
8. Combining the Optimized Yeast Cytosine Deaminase Protein Fragment Complementation Assay and an In Vitro Cdk1 Targeting Assay to Study the Regulation of the γ-Tubulin Complex.
P.H.Ear, J.Kowarzyk, M.J.Booth, D.Abd-Rabbo, K.Shulist, C.Hall, J.Vogel, S.W.Michnick
Cell Cycle Oscillators: Methods and Protocols, 1342, 237 (2016)
DOI:10.1007/978-1-4939-2957-3_14

2015
7. Chemical methods for decoding cytosine modifications in DNA.
M.J.Booth, E.Raiber, S.Balasubramanian
Chemical Reviews, 115 (6), 2240-2254 (2015)
DOI:10.1021/cr5002904

2014
6. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution.
M.J.Booth, E.Raiber, S.Balasubramanian
Nature Chemistry, 6 (5), 435-440 (2014)
DOI:10.1038/nchem.1893

2013
5. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation.
I.M.Iurlaro, G.Ficz, D.Oxley, E.Raiber, M.Bachman, M.J.Booth, S.Andrews, S.Balasubramanian, W.Reik
Genome Biology, 14 (10), R119 (2013)
DOI:10.1186/gb-2013-14-10-r119

4. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine.
M.J.Booth, T.W.Ost, D.Beraldi, N.M.Bell, M.R.Branco, W.Reik, S.Balasubramanian
Nature Protocols, 8 (10), 1841 (2013)
DOI:10.1038/nprot.2013.115

3. Dissection of Cdk1-cyclin complexes in vivo.
P.H.Ear, M.J.Booth, D.Chen, C.Hall, J.K.Moreno, J.Vogel, S.W.Michnick
PNAS, 110 (39), 15716 (2013)
DOI:10.1073/pnas.1305420110

2012
2. Genome-wide distribution of 5-formylcytosine in ES cells is associated with transcription and depends on thymine DNA glycosylase.
E.Raiber, D.Beraldi, G.Ficz, H.Burgess, M.R.Branco, P.Murat, D.Oxley, M.J.Booth, W.Reik, S.Balasubramanian
Genome Biology, 13:R69 (2012)
DOI:10.1186/gb-2012-13-8-r69

1. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution.
M.J.Booth, M.R.Branco, G.Ficz, D.Oxley, F.Krueger, W.Reik, S.Balasubramanian
Science, 336 (6083), 934 (2012)
DOI:10.1126/science.1220671
